|
In the case of von Neumann's design for a self-reproducing automaton, it is clear that all four subcomponents (i.e. A, B, C and D) are very explicitly encoded on the tape φ[A + B + C + D]; the environment in which the automaton exists implicitly encodes only very low-level actions in the form of the local transition rules of individual cells.
The analysis of self-reproducing programs in Tierra-like systems above suggests that in these systems, B and D are explicitly encoded on the tape φ[B + D], but A and C are implicitly encoded in the environment (the operating system). Notice that with this design the 'genetic code' which maps the genotype φ[B + D] to the phenotype [B + D] cannot itself evolve, because the interpretation automaton A is not encoded on the tape.
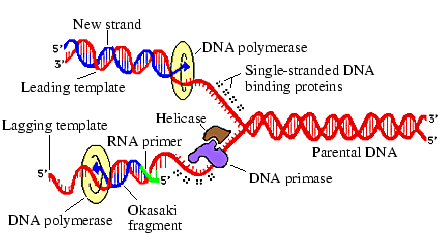
It is interesting to ask how the process of biological cell division fits into this picture. To a first approximation, the information contained in the genome can be thought of as the tape which encodes all of the inheritable information of the cell.7.13 The interpretation of the DNA involves its transcription to messenger RNA, and the translation by ribosomes of this mRNA into proteins. All of the molecular components involved in this process are ultimately derived from information contained in the genome.7.14 The genome therefore contains an explicit encoding of the interpretation machinery A.7.15 The process by which the tape (the genome) is copied is much more complicated than in the artificial systems we have considered up to now. An illustration of the important processes involved in the replication of a DNA double-helix is shown in Figure 7.2.
Many enzymes are involved in the replication process. Replication begins when a topoisomerase enzyme initiates the unwinding of the normally supercoiled DNA. Once this is accomplished, another enzyme, helicase, unwinds the double strands of the helix. DNA polymerase then travels down the single strands of DNA, recruiting free `raw materials' (deoxy-nucleotide-triphosphates, or dNTP's) to hydrogen bond with their appropriate complementary bases on the single strand, and forming a covalent bond with the previous nucleotide on the newly emerging second strand. The formation of these covalent bonds on the new strand is catalysed by the DNA polymerase, and also by another enzyme, ligase. The DNA polymerase can only build up the new second strand once this strand has been initiated by an RNA primer synthesised by the enzyme DNA primase. DNA polymerase also plays a proofreading function, replacing any incorrectly inserted bases as the new strand grows. A number of other enzymes are also involved in the process, and in maintaining the stability of the original DNA as replication proceeds.
Now, despite the immense complexity of this process, the fundamental principle by which the DNA strands are copied is the complementary base-pairing of the dNTP's. Many of the enzymes are required to get the original double-stranded DNA into a state in which it can be replicated, and can therefore be viewed as part of the process that controls when replication takes place (i.e. automaton C in von Neumann's architecture). The proofreading aspects of DNA polymerase improve the fidelity of the copy, but are not fundamental to the replication procedure per se (although these aspects do have important evolutionary implications). Of course, some of the enzymatic functions must be properly viewed as essential to the copying process itself, particularly those enzymes which join the individual segments of the new strand together to form a single continuous molecule. Therefore, some of the copying process proper is governed by enzymes and therefore ultimately encoded upon the DNA itself, but a major element of the process (complementary base-pairing) is determined by the inherent bonding affinities of the molecules, and is therefore implicit in the laws of physics and chemistry.
We might therefore say that the genome of a biological cell corresponds to a tape φ[A + b + C + D] in von Neumann's architecture, where the lower-case b denotes the fact that the fundamental copying process is implicit in the environment, even though extra machinery is involved and explicitly encoded in the genome.
It is interesting to speculate on what information we might desire to be explicitly encoded on a molecule or other structure which would be suitable for acting as a robust initial seed for an open-ended evolutionary process. I will refer to such a structure as `proto-DNA'. Now, we would like our proto-DNA to be an indefinite hereditary replicator if it is to be such a seed. In other words, it should be able to exist in an unlimited number of configurations which retain the ability to reproduce. If the copying process is encoded on the tape itself, then mutations have the potential to disrupt its ability to be reproduced. It would therefore seem desirable that the copying automaton B of our proto-DNA be largely implicitly encoded in the environment. Note that this would not necessarily prevent a more complicated, and possibly more reliable, explicit copying process B' later evolving from (but still based upon) the simpler implicit process, as indeed seems to have happened during biological evolution.
If the copying procedure for our proto-DNA is implicitly encoded in the environment, however, any configuration of proto-DNA would, all else being equal, be able to reproduce as well as any other. In other words, there would be no basis for preferentially selecting some configurations over others, and therefore no basis for an evolutionary process. Specific configurations of proto-DNA must therefore have some specific properties that are selectively significant. Models of the origin of life commonly presume that these simple phenotypic properties were things such as increased stability of the molecule, simple control of the local environment, catalytic activity, etc. (e.g. [Eigen & Schuster 77], [Cairns-Smith 85], [Szathmáry & Demeter 87]).
At the initial stages of an evolutionary process, however, we would not expect there to be mechanisms for explicitly decoding the proto-DNA; in other words, the interpretation machinery A is implicit. This means that particular configurations of proto-DNA should have some specific phenotypic properties (such as the ability to act as catalysts) which can be determined directly from their structure rather than having to be explicitly decoded from the genotype. We could therefore regard the proto-DNA as merely , φ[D] meaning that particular configurations have particular phenotypes associated with them, which are (a) not related to the process of self-reproduction per se, and (b) do not require to be decoded by an explicit interpretation automaton A. Regarding the kinds of simple phenotypes that we might wish to be available to our proto-DNA, some possibilities are suggested by the origin-of-life models mentioned previously, but in general the options seem endless. Graham Cairns-Smith observes:
``It is almost too easy to imagine possible uses for phenotype structures--because the specification for an effective phenotype is so sloppy. A phenotype has to make life easier or less dangerous for the genes that (in part) brought it into existence. There are no rules laid down as to how this should be done.'' [Cairns-Smith 85] (p.106).If more complicated phenotypes are to arise later on in the evolutionary process, however, we require that the proto-DNA at least has the potential for explicit interpretation machinery A' and control machinery C' to become associated with it. This would involve some form of specific reaction to subsections of information in the proto-DNA, but more work is needed to fully identify how this potential for explicit interpretation might be assured.